All published articles of this journal are available on ScienceDirect.
Cardiovascular Magnetic Resonance Imaging in Experimental Models
Abstract
Cardiovascular magnetic resonance (CMR) imaging is the modality of choice for clinical studies of the heart and vasculature, offering detailed images of both structure and function with high temporal resolution.
Small animals are increasingly used for genetic and translational research, in conjunction with models of common pathologies such as myocardial infarction. In all cases, effective methods for characterising a wide range of functional and anatomical parameters are crucial for robust studies.
CMR is the gold-standard for the non-invasive examination of these models, although physiological differences, such as rapid heart rate, make this a greater challenge than conventional clinical imaging. However, with the help of specialised magnetic resonance (MR) systems, novel gating strategies and optimised pulse sequences, high-quality images can be obtained in these animals despite their small size.
In this review, we provide an overview of the principal CMR techniques for small animals for example cine, angiography and perfusion imaging, which can provide measures such as ejection fraction, vessel anatomy and local blood flow, respectively. In combination with MR contrast agents, regional dysfunction in the heart can also be identified and assessed. We also discuss optimal methods for analysing CMR data, particularly the use of semi-automated tools for parameter measurement to reduce analysis time. Finally, we describe current and emerging methods for imaging the developing heart, aiding characterisation of congenital cardiovascular defects.
Advanced small animal CMR now offers an unparalleled range of cardiovascular assessments. Employing these methods should allow new insights into the structural, functional and molecular basis of the cardiovascular system.
INTRODUCTION
Cardiovascular magnetic resonance (CMR) imaging has emerged as one of the most valuable tools for the diagnosis of heart disease. CMR is the only modality able to assess cardiac anatomy, function, viability, perfusion and metabolism within a single non-invasive examination [1, 2]. Improvements in magnetic resonance (MR) scanner performance and hardware have rapidly increased the reliability and accuracy of clinical CMR. More recently, the introduction of parallel imaging and dynamic undersampling techniques has led to substantial accelerations in image acquisition to the extent that real time CMR is now possible. Such technology has opened up the opportunity to use magnetic resonance imaging (MRI) to guide interventional surgical procedures, in addition to further improving the spatial and temporal coverage in the most demanding techniques like first pass perfusion imaging.
CMR in small animals, however, still remains a significant technical challenge, primarily due to the spatial and temporal requirements. Applying MRI to a mouse heart, which can be less than one centimetre in length and beating in excess of 600 times per minute, requires specialist expertise and equipment. Nevertheless, CMR still offers the most accurate means to measure cardiac function and provide unrivalled images of heart anatomy in vivo, allowing for serial non-invasive assessment of animals in cardiovascular research [3, 4]. In this review we aim to provide an overview of the current CMR techniques routinely applied to experimental models of cardiovascular disease in adult mice and rats, and additionally give an insight to some of the emerging techniques that may enable a greater range of possible CMR studies.
1. TECHNICAL CONSIDERATIONS
1.1. MR Acquisition and Hardware
CMR has transformed the way cardiac function can be assessed in small animals, where left ventricular (LV) structure, function, perfusion and viability can all be evaluated within one scanning session, while maintaining a high degree of accuracy and avoiding the harmful effects of ionising radiation and toxic contrast agents. Most cardiac MRI experiments in small animals are performed using dedicated research scanners with a relatively small bore size (typically 20-30cm) compared to clinical systems. The strength of the magnetic field used in MR scanners determines the amount of signal available from a given scan duration and for this reason field strengths of between 4.7 and 9.4T are commonly used for imaging small animals [5], although systems of up to 21.1T are now being installed [6]. High-field scanners are also equipped with high performance gradient systems which are needed to achieve the necessary image resolution. Nevertheless, small animal imaging has been successfully performed on standard clinical systems using dedicated radio frequency (RF) coils, but inevitably the images produced do suffer from reduced signal associated with lower field strengths [7].
1.2. Physiological Monitoring and Gating
Unlike MRI of other anatomy, where the target organs are usually (or close to) stationary, the rapid movement of a beating heart poses a particular challenge in cardiac imaging. Motion is a fundamental vulnerability of MRI, and typically manifests as a ghosting artefact which can severely degrade image quality. While the normal range of resting heart rate in humans is 60-100 beats per minute (bpm), it can reach in excess of 600 bpm in small animals. Therefore, a technique for compensating the effect of cardiac motion is essential for successful cardiac MRI. Prospective electrocardiogram (ECG)-gating is one of the most established techniques used in both human and small animals. The principle of ECG gating is to use the ECG waveform, typically the R-wave peak, to trigger data acquisition, and thus synchronising images to a particular stage in the cardiac cycle. The result is an image of a “frozen” heart, built up from many cardiac cycles. The validity of this technique is therefore limited by the assumption that there is negligible variation between heartbeats, which is true in most cases except in arrhythmias. Although the use of ECG-gating is well-established clinically, and can produce excellent results, specialised ECG equipment is required for use in small animals. These systems are designed to cope with the rapid heart rates, smaller ECG signals and increased electrical interference generated by the large gradient switching required to produce high resolution images.
Respiration is another source of undesired motion; the thoracic cavity and mediastinal organs such as the heart are displaced significantly during respiration. One solution is the use of MR navigator-echoes acquired during image acquisition to track the position of the diaphragm during respiration, allowing the subject to breathe freely throughout. However, with small animals it is most common to track respiratory movements via a neonatal apnoea pressure sensor placed over the chest or an inductive loop [8]. The combined cardiac and respiratory signals provide the essential gating information needed to synchronise image acquisition and to also provide remote physiological monitoring of anaesthetised animals within the scanner.
1.3. Blood Flow Effects
Even with careful gating strategies, images can be degraded by fast flowing ventricular blood at certain parts of the cardiac cycle. Images can suffer from local signal losses or even more detrimental flow artefacts depending upon the speed of data acquisition. Fast MRI acquisition sequences, such as spoiled gradient echo (GRE), are required to image the heart with so-called bright blood contrast (as shown in Fig. 1). An alternative way of suppressing flow-induced artefact is black-blood imaging, where the blood appears dark, which typically involves a double inversion preparation pulse prior to acquisition. This technique is particularly useful for imaging structural abnormalities of the ventricles and pericardium.

Example of bright blood cine images of a rat heart at 9.4T. Standard orientations consisting of two and four chamber long-axis and short-axis views are acquired. The upper row shows corresponding images for end diastolic (ED) time frame and the lower row end systolic (ES) time point. la: left atrium, mv: mitral valve, lv: left ventricle, ra; right atrium, rv: right ventricle, lvw: left ventricular wall, rvw: right ventricular wall, pap: papillary muscle. The scale bar represents 1 cm.
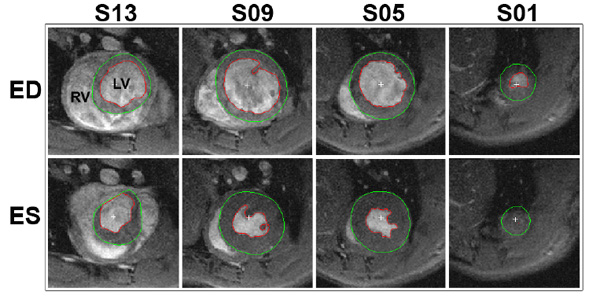
Segmentation of a typical cine stack of a rat heart. The red line delineates the endocardial border, whilst the green line marks the epicardial border. The top and bottom rows depict the end diastolic (ED) and end systolic (ES) time frames, respectively. The columns show different slices where the most apical slice included is labelled as slice 1 (S01). Parameters; in-plane resolution: 200x200 µm, slice thickness 1 mm, RV: right ventricle, LV: left ventricle.
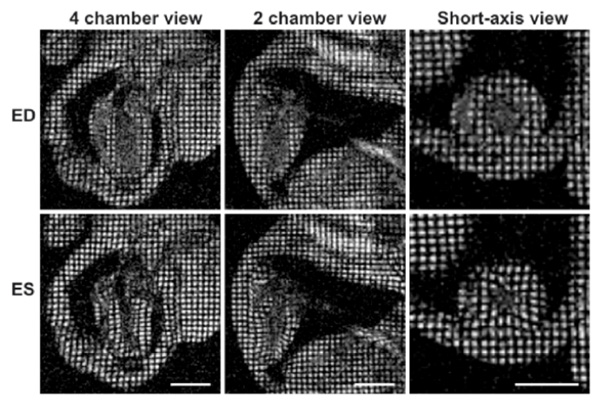
Myocardial tagging in the mouse. Four chamber, two chamber and short-axis views of a mouse heart at end diastole are shown in the top row, while end systolic frames are depicted in the lower row. The movement of the tagged myocardial tissue can be tracked to calculate myocardial strain. The scale bar represents 5 mm.
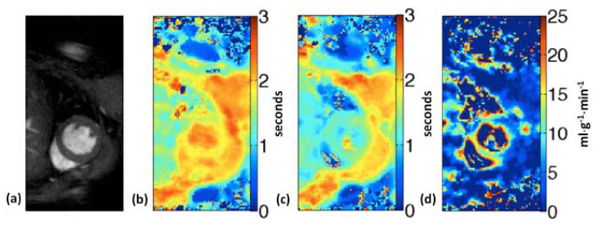
ASL perfusion imaging in the mouse. Short-axis image (a), T1 maps from global inversion (b) and slice-selective inversion (c) are used to generate the perfusion map (d) obtained from a mouse at baseline.
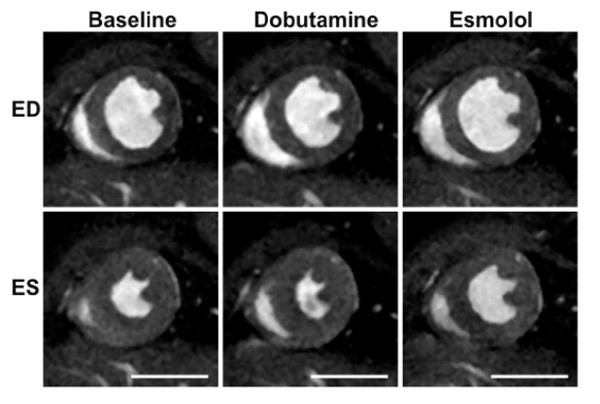
Assessment of pharmacological agents on cardiac function in a normal mouse. Baseline images (first column) acquired prior to an infusion of dobutamine (middle column). In addition, the effect of the beta blocker esmolol (right column) is subsequently shown to counteract the increased contractility caused by the cardiac stress agent dobutamine. ED; end diastolic, ES; end systolic time frames. The scale bar represents 5 mm.

Late gadolinium enhancement MRI in a rat 2 hours following surgery to induce myocardial infarction. A series of consecutive shortaxis images reveal a large area of non-viable myocardium extending down to the apex. Imaging was performed ~10 minutes following injection of Gd-DTPA. From reference [61].
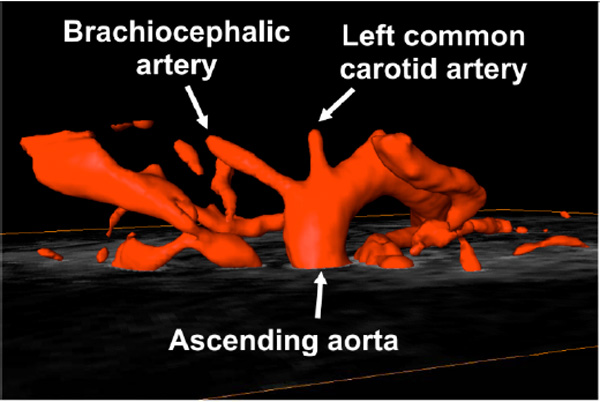
In vivo magnetic resonance angiography of an adult mouse showing the major vessels superior to the base of the heart. TOF techniques provide high contrast between flowing blood and surrounding tissue, image data can be reconstructed using maximum intensity projection to give 3D renders of vessel geometry.

Permanent occlusion model of myocardial ischaemia in the mouse. Late gadolinium images shown here are acquired 48hrs after myocardial infarction (MI) surgery with 3 consecutive short-axis slices (a), and a 4-chamber long-axis view (b), revealing myocardial ischaemic region and the early stages of wall thinning (figures from reference [61]).
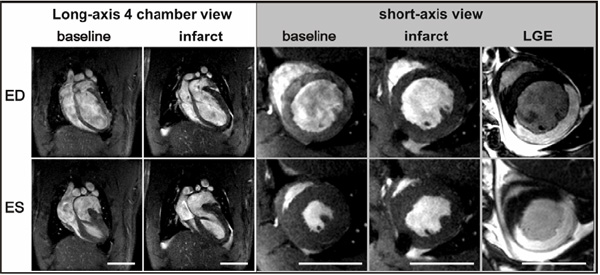
Example of reperfused MI in a rat. Images of the infarcted heart were acquired two hours after reperfusion of the occluded left anterior descending coronary artery. The size of the infarcted myocardium can be roughly estimated via reduced contractility on short axis views. Late gadolinium enhancement (LGE) (far right) can be used for accurate assessment of the infarct size. The size changes associated with cell death are exploited with this method and can be seen as enhancement of the myocardium. ED; end diastolic and ES; end systolic time points. The scale bar represents 1 cm.

Short axis view of a rat heart: mid-ventricular slice at end systolic time point. The arrow points to the hypointense region where stem cells were injected one week prior to image acquisiton into the border region of an infarct. T2* weighted spoiled GRE sequence used.

Outline of the high-throughput embryo imaging process. From left to right: multiple embryos can be embedded into a centrifuge tube and imaged overnight (52µm isotropic resolution). A 3D volume is then produced which can then be viewed at any angle. A sagittal slice from a single embryo shows the quality of anatomical detail in resulting images.

Normal mouse embryo heart anatomy at 15.5 days post coitum (dpc) as seen by µMRI. a-d show successive axial slices through the thorax at the levels indicated. Key: Tr=Trachea, Th=Thalamus, AA=Ascending Aorta, PA=Pulmonary Artery, L.SVC=Left Superior Vena Cava, LA=Left Atrium, RA=Right Atrium, LV=Left Ventricle, RV=Right Ventricle. Note: dimension of heart is approximately 2 mm in diameter.

Axial slices through Chd7+/+ and Chd7+/- embryos as part of a high throughput MRI screen. (a) Image showing a wild-type embryo with an intact ventricular septum. (b) image from an Chd7+/- embryo showing a ventricular septal defect (VSD) which was confirmed on later H&E histology (c) [134]. Volume rendering (d) of the same Chd7+/- heart and great vessels allows visualisation of the VSD (red triangle) in 3 dimensions.
1.4. Pulse Sequences
In clinical cardiac MRI nearly all anatomical and functional cardiac imaging is performed using either a segmented fast spoiled GRE sequence, or more recently, the steady-state free precession (SSFP) technique has become the gold standard. Segmented spoiled GRE images are primarily T1-weighted and as a result the bright blood signal depends on fresh unsaturated (i.e. as yet unperturbed by RF energy) blood flowing through-plane to provide the contrast with the more saturated myocardium. SSFP on the other hand is somewhat independent to blood flow with image contrast dependent on the ratio T2/T1 and as a result provides excellent blood-myocardium contrast even with slow moving blood [9]. In addition to the improved contrast, SSFP is also one of the most signal to noise ratio (SNR) efficient sequences, providing greater signal in less time than spoiled GRE images. Unfortunately, SSFP images suffer from banding artefacts which are related to how short the repetition time can be made and the homogeneity of the magnetic field. As field inhomogeneity is directly related to field strength, it is extremely difficult to use SSFP for small animals at high fields. Thus, the primary sequence for assessing cardiac function in small animals is the original spoiled GRE used in the early days of human cardiac imaging. Following R-wave peak detection, a set number of echoes or k-space lines of data are acquired to make up the multiple frames of a cine image set. The process is repeated for many heart beats, typically 128 or 256 times, until all the data needed to reconstruct a full cine image stack are acquired, creating a movie (or cine) of the beating heart. Standard image orientations of the heart consisting typically of a 2- and 4-chamber long axis view and, in order to calculate global function, a complete stack (from apex to base) of short axis images are acquired (see Fig. 1).
1.5. Image Analysis
For almost all the animal models for which CMR is used, data analysis will be necessary to calculate functional parameters of the heart. Such parameters may be estimated for comparison at a single time point or for repeated measurements in order to study disease progression or treatment response. A typical CMR study will assess the standard global functional and morphological parameters of the heart, including ejection fraction (EF), end-diastolic volume (EDV), end-systolic volume (ESV), stroke volume (SV), and left ventricular mass (LVM). Additional parameters such as myocardial perfusion, infarct size, myocardial strain, peak ejection and filling rates or the effect of agents such as dobutamine may also be assessed.
Central to any CMR study is the analysis of cine data for the standard functional parameters. These are commonly calculated from a series of cine images taken from the apex to the base of the heart, which require accurate detection of the epicardial and endocardial borders (see Fig. 2). A clear set of rules for the inclusion of slices, maximum heart rate variability between slices, the handling of papillary muscles and the definition of the valve planes should be adapted to ensure accuracy, as well as low inter- and intra-observer variability. In addition, data should always be randomised and blinded before analysis to avoid observer bias.
In many published studies, papillary muscles are often not segmented as myocardium and treated as part of the blood pool. However, it can be argued that papillary muscles should be segmented as myocardium since they account for a sizable fraction of the blood-pool volume at end-diastole (up to 10%) [10]. This approach is commonly adopted due to its simplicity, as the geometry of the blood pool is made much simpler (i.e. circular) by omitting the papillary muscles, thus making segmentation and measurement more time efficient. Nevertheless, the potential effects of omitting the papillary muscles on measurement accuracy should be considered during analysis, especially with estimation of ESV in which the LV blood-pool volume is lowest.
Manual segmentation of CMR data is commonly used and yields low inter- and intra-observer variability [3, 11]. The accuracy of heart volume measurements via MRI has been validated by comparing calculated myocardial mass and ex vivo measurements, showing a high correlation between both methods [3, 12, 13].
The increasing demand for CMR and the time consuming nature of manual segmentation has lead to development of commercially available semi-automatic segmentation methods. These may offer a considerable reduction in analysis time while showing similar inter- and intra-observer variability [11, 14]. Such segmentation programs use different approaches from simple thresholding to more advanced deformable models for volume estimation and generally include long axis views to improve valve definition accuracy.
2. TECHNIQUES FOR ASSESSING CARDIAC PHYSIOLOGY
2.1. Myocardial Mass
Myocardial mass, or left ventricular (LV) mass in particular, is an important index for the assessment of cardiac functional status for a variety of clinical conditions such as cardiomyopathy, hypertension, and valvular disease. MRI can be used to quantify myocardial mass by acquiring a volume dataset of the heart. A delineation of the epicardial and endocardial contours of the left ventricle is performed, after which the segmented volume is multiplied by the myocardial specific gravity (1.05 g/cm3) to calculate the LV mass. Accurate in vivo quantification of myocardial mass using MRI has been demonstrated and validated in the mouse and rat [4, 15, 16]. Clinically, MRI has been shown to be more accurate and reproducible than M-mode, 2D and 3D echocardiography [17-19].
2.2. Wall Motion
Global and local myocardial motion can be assessed by myocardial tagging strain analysis, using sequences such as spatial modulation of magnetisation (SPAMM) or phase contrast MRI [20]. In SPAMM, spatially selective RF pulses are applied to pre-saturate thin planes of myocardium perpendicular to the imaging plane at end-diastole (see Fig. 3), which are then used as markers for regional myocardial movements [21, 22]. MRI tagging can be applied in two or three dimensions to assess radial, circumferential and longitudinal shortening [23, 24]. The pre-saturated thin planes, or tags, appear dark on the image and persist throughout a cardiac cycle. During myocardial contraction, these tags are displaced in accordance to their underlying myocardial motion. The degrees of displacement are then used to evaluate myocardial contractility using tag detection and displacement-estimation algorithms [25, 26]. Myocardial displacement is calculated and reconstructed in three dimensions to provide detailed strain maps of the tagged myocardium in relation to time and space [24]. Although an accurate assessment of three-dimensional LV function can be obtained with this technique, data analysis is slow and can require significant user input [27].
Faster data processing and image acquisition techniques such as; harmonic phase, displacement encoding with stimulated echoes [28], and fast strain-encoded MRI [29] are emerging. Harmonic phase MRI utilises the phase component of images acquired with SPAMM, which contains an array of harmonic (or spectral) peaks in the Fourier domain. A single off-centred peak contains motion information, where the phase of a complex image (inverse Fourier transform) of such a peak is linearly related to the true motion [30]. By isolating these peaks, the motion of the underlying tissue could be tracked automatically with great accuracy [31].
Myocardial tagging has been validated in a canine model with sonomicrometry [32], and was used to evaluate myocardial strain during local ischaemia in dogs [30, 33] and after myocardial infarction in rats [27]. It has also been applied to characterise regional myocardial function in the mouse [23] and to demonstrate that left ventricular torsion is equal in mice and humans [34].
2.3. Perfusion
Perfusion measurement plays an important role in early detection of occlusive coronary heart disease, as well as determining the severity of occlusion and viability of myocardium [35]. A characteristic feature of occlusive coronary heart disease is the reduction in myocardial flow reserve, which can be detected before clinical symptoms or LV dysfunction become evident [36]. Currently, there are two main techniques for perfusion studies: Arterial spin labelling (ASL) and contrast-enhanced first-pass perfusion imaging.
The first-pass perfusion technique tracks the transit of blood by using gadolinium-based contrast agents; gadolinium is paramagnetic and shortens T1 relaxation, thus giving higher signal intensity in T1-weighted images. During a first-pass perfusion scan contrast agent is injected peripherally, which then passes through the venous system into the heart chambers. A fast (single-shot) T1-weighted sequence is used to image the myocardium as the contrast agent reaches the coronary arteries and perfuses through, with enhancement peaking approximately 10 heartbeats after passing the LV chamber [37]. Signal intensity correlates with contrast concentration, and hypo-enhanced areas indicate reduced or no tissue perfusion. The technique is well-established clinically and is the most practical method for evaluating myocardial perfusion [37], however it remains a significant challenge for use in small animals. In order to capture the arrival of the first-pass of contrast agent, complete image frames need to be acquired in a relatively motion-free portion of the cardiac cycle, so for mice this could be in the order of 10-30 ms. In addition, reproducible bolus injections are hard to achieve in small animals, making accurate quantitative perfusion studies extremely difficult to perform [5, 38].
Alternatively, arterial spin labeling (ASL) may be used to non-invasively quantify perfusion. ASL perfusion measurements are generated by the comparison of two T1 maps: i) using a global inversion recovery and ii) with a slice-selective inversion recovery for a slab slightly larger than the imaging slice. In the slice-selective case, spins (proton signal) that enter the imaging slice due to perfusion are not inverted, causing an apparent acceleration in T1 recovery. An assessment of the difference between the global and slice-selective T1 maps provides a quantification of the blood perfusion in the imaging slice. Fig. (4) displays T1 maps generated by global and slice-selective inversion and a perfusion map for a mouse at baseline under general anaesthesia.
This technique has been adapted for use in the myocardium of mice and rats in a limited number of studies (sequences are presented in [39-41]). Blood flow values have been found to be comparable with microsphere measurements [41-43], although low SNR and speed are the main limitation of this technique. To date, ASL has been used to assess perfusion in a diverse range of applications, including myocardial infarction models [41], nitric oxide synthase knockout mice [44], dobutamine stress tests [40], vasodilatation tests and effects of a variety of anaesthetics [44-46].
Another technique, widely used in functional neuroimaging, is blood oxygen level dependent (BOLD) imaging, and has been applied to assess myocardial perfusion [47]. Traditional BOLD imaging is based on T2*-weighted sequences as previous research in the brain has shown T2* to be more sensitive than T2 to changes in oxygen saturation [48]. However, T2*-based BOLD cardiac imaging poses significant problems due to local field inhomogeneities, exacerbated by the susceptibility of a GRE sequence with long echo times [49]. Alternatively, T2 could be used, but is less sensitive to oxygen saturation changes. By using a T2-prepared SSFP BOLD sequence, Shea et al. demonstrated a good correlation between changes in signal intensity and myocardial blood flow [49].
2.4. Viability
Myocardial viability is defined as the presence of living (or viable) myocytes, and is an important index in assessing the extent of post-ischaemic injury. It is important to note that a portion of myocytes might have impaired contractility after an ischaemic assault, possibly due to reduced perfusion, but are still alive. These cells are known as hibernating myocytes and their functionality is potentially fully recoverable when normal myocardial blood flow is restored. Two frequently used clinical MRI techniques, dobutamine stress and late gadolinium enhancement, offer the means to differentiate between hibernating and non-viable myocytes.
In dobutamine stress MRI, a series of cine images are acquired while increasing doses of dobutamine, an inotropic agent, are infused into the subject (see Fig. 5). Any akinetic or dyskinetic myocardial segments that are viable (ie. contain hibernating myocytes) would show an improvement in contractility after administration of dobutamine. This technique has been shown to adequately predict improvement in LV function in patients after revascularisation [50], and was employed to investigate acute and chronic cardiac remodelling after MI in mice and rats [51, 52]. Myocardial tagging can be incorporated into dobutamine stress MRI and has demonstrated superior accuracy over studies without tagging [53], and produced similar accuracy to dobutamine stress echocardiography (DSE) [54]. Currently, there are still only a limited number of experimental studies that have utilised dobutamine stress MRI in small animals [16, 44, 51, 55].
The other approach for assessing myocardial viability is delayed enhancement MRI or late gadolinium enhancement (LGE) which involves administering a contrast agent to directly image non-viable myocardium (see Fig. 6). In this method, an extracellular contrast agent (usually Gd-DTPA) is injected into the subject. On first-pass images, normal myocardium with normal perfusion would be enhanced by the contrast agent, whereas infarcted regions would appear hypoenhanced due to reduced or absent perfusion. However, the hypoenhanced region would gradually become hyperenhanced as a consequence of increased retention of contrast agent, which is thought to be caused by increased extracellular matrix within collagenous scar, and delayed washout due to reduced capillary density [56]. This method is well-validated clinically for predicting the distribution of infarcted myocardium [57] and for excluding irreversible damage where no delayed enhancement is observed [58, 59]. It is also well-validated in a mouse occlusion/ reperfusion model with 2,3,5-Triphenyltetrazolium chloride staining [60], but results are varied in the rat model [5].
Manganese-enhanced MRI (MEMRI) is an alternative technique used in experimental cardiac studies. Similar to Gd agents, Mn2+ is also paramagnetic and primarily acts by reducing T1 of surrounding tissue to cause signal enhancement on MR images. However, its strength in small animal studies is its use as a molecular agent, since Mn2+ ions have physical properties similar to calcium ions, and therefore taken up by excitable cells such as neurons and cardiomyocytes where Ca2+ influx readily occurs. In cardiomyocytes in particular, Mn2+ is able to enter the cell through voltage-gated calcium channels. However, in contrast to Ca2+, Mn2+ is retained after uptake, which can lead to cardiotoxicity.
The key use of MEMRI in cardiac imaging is to produce a signal enhancement on MRI that relates to cardiac function. In the mouse heart, MEMRI has been shown to be sensitive to inotropic changes [62]. Administering Mn2+ by intravenous infusion, forming a stable plasma concentration allows baseline signal intensity measurement to be taken. After administration of a inotropic agent, such as dobutamine, MRI signal can be seen to increase in more active areas of myocardium. Similarly, in animal models of myocardial infarction, Mn2+ may also be used to delineate between infarcted and healthy myocardium on the basis that uptake of Mn2+ is greater in regions of functioning tissue where viable cardiomyocytes are abundant.
2.5. Angiography
Magnetic resonance angiography (MRA) in small animals is most commonly performed using the time-of-flight (TOF) approach [63-65]. The technique relies on the same contrast mechanism utilised in bright blood imaging, but typically 3D images are collected from multiple acquisitions of overlapping thick slabs rather than 2D slices. Vascular anatomy can then be visualised with rendered maximum intensity projection 3D volumes (see Fig. 7). Most applications of small animal MRA have been limited to cerebro-vascular anatomy, applying the technique to the vessels surrounding the heart is particularly challenging. However, there have been reports of using TOF MRA to visualise even the coronary arteries in the living mouse [66]. Low-doses of gadolinium contrast agents can be used to further enhance the TOF effect and improve image quality. They can be used to reduce the overall acquisition time which can be particularly long when cardiac and respiratory gating is used, which is essential for visualising the smaller vessels located close to the heart [67].
2.6. Vessel Wall and Plaque Imaging
Vessel wall imaging in small animals remains challenging due to the extremely high resolution required. Ohlstein et al [68] and Chandra et al [69] performed in vivo (at 4.7T) and ex vivo (at 9.4T) carotid artery imaging in rats. In these ex vivo studies, the various layers of the arterial wall were clearly defined. In vivo vessel wall imaging of the mouse common carotid artery was performed in another study [70] by using a composite image-reconstruction method, where T1, T2 and PD-weighted images were assigned a separate colour channel, and combined to form a 24-bit colour image. The resultant contrast made the vessel wall distinguishable, but not sufficient for accurate measurements. In another study, the mouse coronary arteries and the four valves were successfully imaged in vivo, without the aid of contrast agents [66]. Vessel wall and plaque imaging have also been performed in other knockout mouse models [71-74]. Targeted contrast agents like SPIOs (super-paramagnetic iron oxide particles) conjugated to antibodies can be used to image specific molecular processes involved in disease progression [75].
2.7. Myocardial Micro- and Macro-Structure
Diffusion tensor imaging (DTI) has the potential of characterising the remodelling of cardiac fibre structure after infarction in rats, and is based on the theory that water diffusion will be the greatest parallel to the myocardial fibre orientation, and therefore the primary eigenvector of the diffusion tensor coincides with the local fibre orientation. By comparing DTI and conventional histological results, recent studies using dogs, goats and rabbits [76-78] have demonstrated a good correlation between MRI and histological fibre angle measurements, thereby validating the DTI approach for complete characterisation of myofibre architecture. MR-microscopy and diffusion imaging have also been performed on isolated and fixed rat hearts [79-82]. DTI may potentially be a valuable tool for defining structural remodelling in diseased myocardium at cellular and tissue levels.
2.8. Metabolic Characterisation
MR spectroscopy is a technique which can non-invasively measure the relative concentrations of molecules. It can be used to characterise the concentrations of high energy phosphorous compounds involved in cardiac energy metabolism for a selected region of the myocardium. Phosphorus molecules of interest are adenosine triphosphate (ATP), phosphocreatine (PCr) and inorganic phosphates (Pi). Myocardial ischaemia leads to a significant reduction in the PCr/Pi ratio, as well as a reduction in the PCr/ATP ratio. 31P MR spectroscopy has been demonstrated to be feasible for measurements of the PCr/ATP ratio in isolated mice hearts [83, 84]. Concentration of intracellular sodium is another indicator for myocardial ischaemia, and could be distinguished from extracellular sodium using 23Na MRI with chemical shift imaging [85]. Cardiac metabolic imaging of other nuclear magnetic resonance-visible nuclei such as 13C and 19F have also been investigated, but experience in this field is still limited [86, 87] and is particularly challenging to implement successfully.
3. EXPERIMENTAL ANIMAL MODELS OF CARDIOVASCULAR DISEASE
3.1. Myocardial Ischaemia
Myocardial ischaemia occurs when there is an imbalance between myocardial oxygen supply and consumption due to reduced coronary blood flow [88], which if not restored promptly would ultimately lead to infarction of myocardium. Clinically, myocardial ischaemia is most commonly caused by thrombotic occlusion of a coronary artery, which is initiated by the rupture of an atherosclerotic plaque. Since this is a spontaneous process, it is difficult to mimic; therefore myocardial ischaemia in most animal models is achieved by occluding the coronary arteries mechanically either by direct ligation or intraluminally, with the former being the most popular technique in small animals including mice and rats, and the latter method largely confined to bigger animals such as dogs and pigs [89]. With the direct ligation method, the animal is anaesthetised and ventilated, after which the heart is exposed typically via a left thoracotomy. The left anterior descending coronary artery (LAD), which supplies the anterolateral region of the left ventricle, is then ligated using sutures and a snare. Successful occlusion is evident by the rapid pallor over the anterolateral region of the left ventricle that is perfused by the LAD, as well as associated ECG changes.
The evolution of ischaemia to infarction and subsequent ventricular remodelling is characterised by the progression of injured myocardium to fibrosis and corresponding wall thinning. Compensatory hypertrophy of the remote healthy myocardium may follow, after which the infarcted heart may then progressively dilate and eventually undergo failure [90]. By using animal models, the site, duration and degree of coronary occlusion can be manipulated to alter the characteristics of the resulting ischaemia and infarction, and therefore allowing the different stages of myocardial infarction to be studied. Different variants of myocardial infarction models will be discussed below.
3.1.1. Ischaemia without Reperfusion (Permanent Occlusion)
In a permanent occlusion model, the LAD artery is ligated permanently with a silk suture and therefore the ischaemic myocardium is not reperfused. As a result, infarction is often more extensive and transmural in nature, and subsequent fibrosis and wall thinning is achieved more rapidly (see Fig. 8). This approach is thus valuable for investigating the later stages of myocardial infarction, such as ventricular remodelling and heart failure [91].
3.1.2. Ischaemia with Reperfusion (Transient Occlusion)
This approach involves a transient period of ischaemia followed by the release of ligation to allow reperfusion of the previously ischaemic myocardium (example shown in Fig. 9). Transient occlusion represents more closely the ischaemia-reperfusion scenario that is found clinically, where patients are revascularised following an acute ischaemic event. Therefore, the ischaemia-reperfusion model is popular amongst interventional studies, including those using statins [92] and stem cell therapy [93]. In addition, the ischaemia- reperfusion model is also valuable for investigating reperfusion injury, a phenomenon believed to contribute to further myocardial damage following reperfusion [94].
3.2. Atherosclerosis and Vascular Injury Models
3.2.1. Atherosclerosis Models
Atherosclerosis, a precursor of myocardial infarction and stroke, is a common inflammatory disease of complex pathogenesis and multiple factors such as thrombosis, hyperlipidaemia and endothelial injury. It is characterised by the accumulation of lipids and fibrous components, the recruitment of monocytes and lymphocytes, and hyperplasia of media, all of which contributing to the subsequent thickening and reduced elasticity of the arterial wall [95]. The rabbit is one of the most investigated animals in atherosclerosis research, since they develop spontaneous or high-lipid diet-induced atherosclerotic plaques [96]. While rodents are naturally resistant to atherosclerosis, several transgenic mouse strains such as the apoprotein-E knockout have been developed to introduce lipid abnormalities and subsequently atherosclerosis [97]; such techniques offer the potential to study the role of isolated metabolic pathways in atherosclerosis.
The stability of an atherosclerotic plaque is an important factor, since vulnerable plaques – characterised by a thin cap and large lipid core – are prone to rupture, where the ensuing thrombosis is often the cause of coronary occlusion [98]. Therefore, the ability to differentiate between stable and vulnerable plaques is of great clinical relevance; however, there is currently no robust biomarker to distinguish between the two due to their similar composition and pathophysiology. Recently, imaging techniques such as intravascular ultrasound and high-resolution MRI have provided some promises in identifying vulnerable lesions in the clinical settings [70, 99].
3.2.2. Endothelial Injury and Restenosis Models
Localised arterial wall and endothelial injury can be produced by mechanical means to investigate restenosis (re-narrowing of the vessel) in animal models. This disease is associated with transluminal angioplasty, a technique widely used for coronary revascularisation following myocardial infarction, where a small balloon is inserted and inflated to expand the occluded artery. The procedure invariably causes injury to arterial wall and in particular endothelial denudation, triggering cell proliferation, intimal thickening and ultimately restenosis that may require repeated treatment. In rats, the common carotid artery is often used to create endothelial injury due to its accessibility and size [100]. Assessment of injury and disease progression in this model have traditionally relied on histopathological analysis with cross-sectional slides [101]; increasingly, standard MRI and MRA techniques as well as the more novel MR microscopy and contrast agents are being used to assess the vessel wall and quantify the degree of restenosis in addition to histology [68, 69, 102, 103].
3.2.3. MR Assessment of Cellular Therapies
The discovery of stem cells and their potential for tissue regeneration led to the hypothesis that regeneration of the adult damaged heart might be possible via stem cell transplantation. It is hoped that stem cells will differentiate into cardiomyocytes or fuse with resident myocytes and functionally integrate if they receive the right clues from their microenvironment. Following this hypothesis many studies in animals [104-112] and a few clinical trials [112-114] have been conducted. Some of the cell transplantation studies in small animals showed considerable improvement in cardiac function while others showed only mild improvements. However, these improvements could not be translated into human trials. It is, therefore, necessary to improve the animal models in order to derive clinically relevant information from them. These studies employed different analysis methods such as MRI, PET and echocardiography which is probably one of the sources for variability.
Another problem with small animal models is the quantification of the amount of cells and their specific localisation. MRI together with stem cells labelled with superparamagnetic iron oxide particles (SPIOs) can be used to address these issues. SPIOs generate local field inhomogeneities when placed into a magnetic field. This causes local hypointensity (darkness) in T2 or T2* weighted MR images (see Fig. 10). The sensitivity of T2* images to SPIOs allows the detection of a few thousand cells or even single cells [115-119]. Several publications indicate that the labelling of stem cells with small amounts of SPIOs does not alter their viability and differentiation potential [120-122]. However, there is always the possibility of functional changes. Therefore, a careful optimisation of the labelling strategy should be performed before it is used in vivo. SPIO labelled cells offer the possibility to confirm the delivery success of a particular strategy and allow the tracking of cell movement over time.
The quantification of delivery success is important to establish if there is a dose dependent relationship between the amount of cells delivered and the functional outcome of a study. However, this has to be done with caution. Firstly, imaging artefacts which cause hypointense regions are frequently observed on cardiac cine images. Secondly, the hypointensity observed from SPIOs is only indirectly coupled to the cells administered in particular over a long time course. SPIOs are frequently taken up by neighbouring cells and macrophages when cells die. As long as those cells and macrophages stay in the same imaging voxel they cannot be differentiated from the transplanted cells. A major limitation of SPIO based cell tracking is the inability to assess cell viability. It might, therefore, be necessary to use an additional labelling strategy which allows assessment of cell viability over time. Depending on the method, it might allow for the combination of high spatial resolution MRI with in vivo cell viability assessment.
4. IMAGING THE DEVELOPING HEART WITH MRI
4.1. Ex Vivo Morphological Phenotyping of Mouse Models
Since the early part of the 20th century, mice have been used widely for genetic investigations [123] and more recently methods have emerged to enable direct manipulation of the mouse genome. The mouse has become the key model for research into human disease. With the announcement of the completion of the first draft of the mouse genome in 2002 [124], a number of large-scale programmes are underway to systematically create transgenic knockout models [125]. The study of the embryos of such mouse mutants is useful to determine effects of genes on development and congenital disease. Given the commonality in molecular pathways across the embryo, insight may be obtained into novel genes involved in the basis of heart development. The study of the effect of genes on mouse embryo development is particularly relevant to the investigation of congenital heart defects (CHDs).
Morphological analysis of embryo anatomy is an important part of phenotype characterisation. Conventionally, this is done ex vivo by microscopic examination with histology. In this method, embryos are ethanol-dehydrated, embedded in paraffin wax and thinly sectioned into 2-8µm slices [126]. This provides high resolution 2D data and sections that may also be stained for gene and protein expression. A recent refinement of this process is episcopic imaging, where the autofluorescence of each histology section can be photographed and combined computationally to generate high resolution 3D volume datasets (typically 1-2µm isotropic resolution) [127, 128]. However, these histological approaches are time-consuming and do not lend themselves readily to rapid screening as only one embryo may be imaged at a time. Non-invasive imaging methods, such as MRI, have the potential to offer large time advantages and are increasingly used as an adjunct to histology in the examination of embryo phenotype. This is particularly important considering the large number of embryos required to characterise a phenotype.
4.2. Non-Destructive Imaging in Ex Vivo Embryo Phenotyping
Due to the generally low-volumes of embryos that may be phenotyped with conventional histology and its destructive effects on tissue morphology, there is interest in utilising non-invasive imaging methods for morphological screening. Microscopic MRI (µMRI) is one such imaging method, allowing assessments of both embryo development [129] and phenotype [130].
The use of high-resolution µMRI for cardiovascular phenotyping was initiated in the 1990s by Smith and colleagues [131], where they developed a technique to image embryo vasculature. Later, Jacobs and colleagues produced high resolution MR images of single embryos [132], with the goal of creating 3D anatomical atlases as an advance over the 2D histology-based atlases [126] commonly used, although with a long scan time per embryo. This work has recently been extended by Petiet et al. who produced a database of embryonic, neonatal and adult mice imaged at a number of developmental time points using their method to characterise heart phenotypes in mid-gestation embryos [129, 130].
4.3. µMRI of Multiple Embryos
Techniques have also been developed to image multiple embryos, in a single acquisition [133]. This method has recently been optimised for cardiac phenotyping in developing embryos [134] and can allow for up to 40 embryos to be imaged in a single 10-hour scan, through the use of a customised imaging coil. In this method, embryos are dissected from the mother and fixed for at least two weeks in a solution of formaldehyde and an MR contrast agent, such as Gd-DTPA. The use of these agents as so-called active stains for ex vivo tissue [135], boosts SNR and enhances image quality when performing µMRI. This SNR improvement is important for embryo imaging as their small size requires very high resolution datasets (< ~50 µm isotropic), which can take days to acquire without active staining. As the agent is given sufficient time to penetrate fully into embryo tissues, tissue T1 relaxation time is reduced, allowing shorter scan-times to be used without affecting SNR and thus image quality. An isotropic 3D dataset is then acquired that may be sliced arbitrarily and viewed in any direction. An outline of the imaging process is shown in Fig. (11). The detailed heart anatomy seen in typical MR images is shown in Fig. (12).
4.4. Phenotyping of the Chd7 Model of Charge Syndrome
Cardiac phenotypes can be identified with the multi embryo µMRI technique. One example is screening of Chd7 heterozygous embryos [134]. CHD7 is a transcription factor implicated in CHARGE syndrome [136] – a condition with an incidence of 1 in 10,000, partly characterised by defects the cardiovascular system. Ventricular septal defects were identified using MRI in heterozygous mice (Fig. 13), as seen in axial images and volume rendering; great vessels are also readily visible allowing phenotypes in these regions to be identified.
4.5. Future Direction of Embryo Phenotyping Using MRI
In vivo imaging in utero with MRI has been reported [137, 138] and is able to show structural images of embryos within the mother. However, the continuous foetal motion requires relatively short scans to be taken to reduce movement artefacts in the image. This places a limit on the achievable resolution (typically ~150um in-plane), signal and contrast to noise. An additional difficulty for the assessment of cardiac function in embryos has been the synchronisation of the image acquisition with the cardiac cycle. Recent developments have enabled the acquisition of 3D cine datasets of the embryo heart in vivo [139]. Manganese, which is increasingly used as an MRI contrast agent in preclinical brain studies for activation and tract-tracing applications, has been applied to the embryo [140]. Its use, in conjunction with in utero MRI, could offer functional information to future embryo heart studies.
A full comprehensive analysis of cardiac phenotype and development in embryos will continue to need a complementary set of imaging modalities ranging from optical microscopy to MRI and ultrasound. The strength of MRI, currently, is its ability to rapidly create non-invasive 3D datasets of multiple embryos without distortion and with good delineation of soft tissue and vascular anatomy. However, the traditional strength of MRI has been its capability to perform longitudinal studies, where changes in an individual animal may be followed over time. With advances in in utero techniques it may soon be possible to carry out a temporal characterisation of heart development from embryo to adult.
5. CONCLUSION
The challenge for cardiac imaging is the accurate assessment of cardiac structure and function in a non-invasive fashion with high temporal and spatial resolution. Ideally, techniques should be widely available, suited to both experimental and clinical conditions, and applicable to a variety of physiological and pathological states. Cardiac MRI techniques have become important tools in the research environment with a steadily increasing number of research facilities implementing CMR.
It is clear that these imaging techniques provide a broad range of measures of cardiac integrity that are related to the functional status of the tissue. One of the major advantages of CMR is its non-invasive nature which lends itself to repeated assessment of cardiac function in the same animal. This allows the tracking of pathological changes over time and as the same animal is used repeatedly increases the statistical power to detect differences. This unique information, in the form of high resolution structural and functional images of the heart, may lead to better understanding of cardiac pathologies and potential treatment strategies. Additionally, using techniques from the clinical setting should help to improve the applicability of experimental findings to human patients.
ACKNOWLEDGMENTS
This work was supported by the British Heart Foundation, Biotechnology and Biological Sciences Research Council and the Engineering and Physical Sciences Research Council.